Jmol Tutorial 2
- Home
- Jmol Tutorial 1
- Jmol Tutorial 2
- Jmol Tutorial 3
- Jmol Commands
- Resources Pages
- PDB Tutorial
- SMART Teams
Symposium
Part 2 - Exploring Insulin Dimers
In tutorial one, we started off by looking at the active monomeric form of insulin in order to understand how insulin forms a trimer of dimers. In this tutorial we are going to look at another PDB file in order to look at a different form of insulin. In tutorial one, we modeled the insulin monomer to look like the figure below:
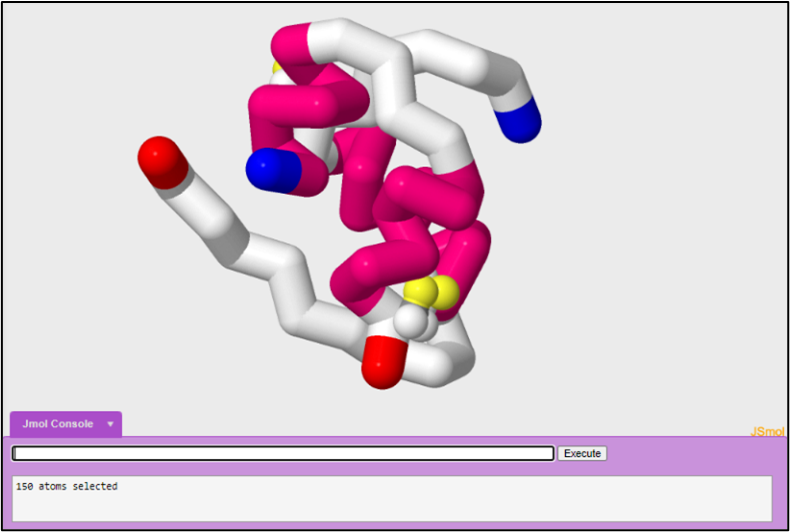
1. PDB file
We will be using 1tyl. When you open the 1tyl file in Jmol it will look like this:
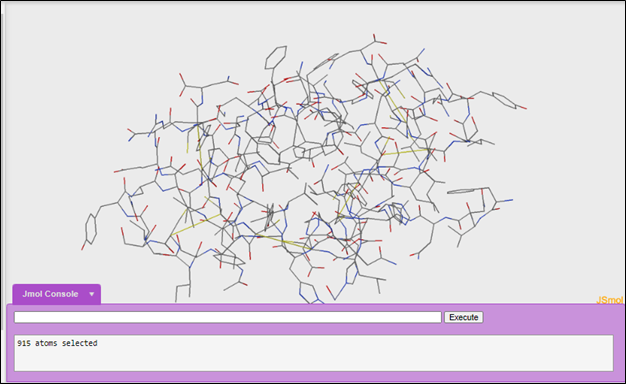
2. Initial exploration of a structure
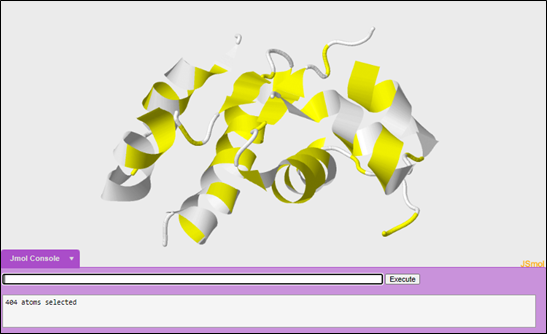
When exploring a structure for the first time, a scientist's goal is to obtain as much information as possible as about the structure of a protein as fast as possible in order to better understand how structure leads to function. You can explore the structure in different display formats. Many scientists will first look at the cartoon display format.
To change your display format into cartoon:
cartoon only
To color the structure to highlight the secondary structures of the protein:
color structure

Questions:
1. How is the form of insulin in the 1tyl file different from the form of insulin in the 2hiu file we looked at in the first tutorial?
2. What information can you get by looking at the cartoon display format?
3. What features of the protein does this model show and what are the model's limitations?
3. Wireframe format
After looking at the cartoon display format, a scientist may switch to look at a different display format like the wireframe format.
To change your display format into wireframe:
wireframe only
To increase the thickness of the wireframe to 0.25 angstroms:
wireframe 0.25
To color the carbon's gray, the hydrogen's white, oxygen's red, nitrogen's blue, and sulfur's yellow:
color cpk
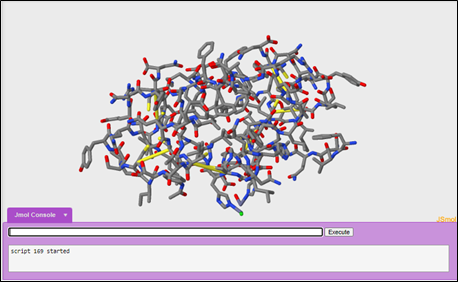
Questions:
4. What information can you get by looking at this display format?
5. What features of the protein does this model show and what are the model's limitations?
Exploring hydrophilic and hydrophobic interactions
In this section we are going to compare and contrast two different 1tyl models in order to draw conclusions about hydrophilic and hydrophobic interactions within the insulin protein. We will make this comparison by highlighting with different colors the hydrophobic portions of the protein in yellow and the hydrophilic portions of the protein in white.
For our first model, we are going to use the cartoon format.
To change your display format to cartoon only:
cartoon only
To color your display format grey:
color grey
To select the hydrophobic portions of the protein:
select hydrophobic
To color the hydrophobic portions yellow:
color yellow
To select the hydrophilic or polar portions of the molecule:
select polar
To color the polar portions of the molecule white:
color white
To make the second molecule, open a new JUDE display window. Then, open another 1tyl molecule.
To change the display to wireframe only:
wireframe only
To increase the thickness to 0.25 angstroms:
wireframe 0.25
To select the hydrophobic portions of the protein and the sidechains:
select hydrophobic and sidechains
The use of and in this command is a boolean operator. This command selects for the hydrophobic portions of the protein and the sidechains. Remember an 'and' boolean operator has to fulfill both conditions.
To color yellow:
color yellow
To select the polar portions of the protein and the sidechains:
select polar and sidechains
The use of and in this command is a boolean operator. This command selects for the hydrophobic portions of the protein and the sidechains. Remember an 'and' boolean operator has to fulfill both conditions.
To color white:
color white
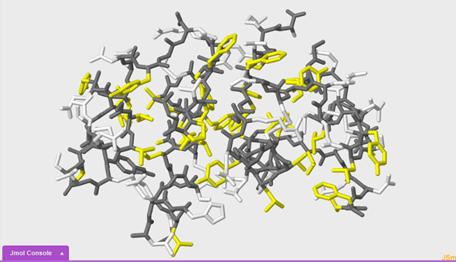
Questions:
6. Compare the two images that you just made side by side. What do you notice?
7. As you observe the model, what questions about insulin structure come to mind?
A deeper look at secondary structure
One advantage to looking at the wireframe format is that you can explore secondary structure in more detail by exploring one alpha helix in the molecule and adding in hydrogen bonds. We are going to explore the alpha helix on chain b.
To make sure your entire molecule is selected:
select all
To change the display to wireframe:
wireframe only
To increase the thickness to 0.25 angstroms:
wireframe 0.25
To color the carbon's gray, the hydrogen's white, oxygen's red, nitrogen's blue, and sulfur's yellow:
color cpk
To only display the helix on chain b:
restrict :b and helix
Notice how the restrict command removes the display of everything except what was restricted. The use of and in this command is a boolean operator. This command selects for chain b and the alpha helix. Remember an 'and' boolean operator has to fulfill both conditions.
To display hydrogen bonds:
calculate hbonds
To set hbond solids:
set hbonds solid
To color hbonds white:
color hbonds white
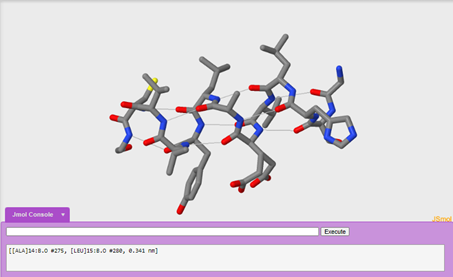
Questions:
8. Which amino acids are contributing to secondary structure? (Hint: if you click on the structure, the console window will tell you what amino acid and atom it is.)
9. What pattern do you see with hydrogen bonding in the alpha helix?
10. How does hydrogen bonding contributes to secondary structure? Be specific!
11. Which bonding interactions occur first: the covalent bonds between amino acids or hydrogen bonding? Why?
12. Try highlighting the hydrophobic sidechains and the polar sidechains with different colors, like we did in the previous section. What do you notice?
13. Based upon your observations from question 12, what questions about insulin structure do you now have?
Comparing secondary and tertiary structure
We are now going to compare secondary and tertiary structure using the wireframe format. You will need to open a new blank JUDE display window. Then, open another 1tyl molecule.
To change the display to wireframe only:
wireframe only
To increase the thickness to 0.25 angstroms:
wireframe 0.25
To hide the sidechains of each amino acid:
hide sidechains
To calculate hbonds:
calculate hbonds
To color hbonds white:
color hbonds white
To turn on disulfide bonds:
ssbonds on
To show ssbonds with backbone only selected:
set ssbonds backbone
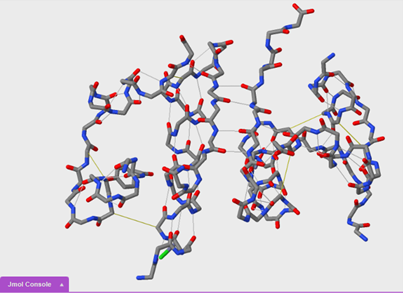
Questions:
14. Based on what you observed, which bonds form first, the hydrogen bonds of an alpha helix or the disulfide bonds between helices? Why?
15. Order the formation of disulfide bonds in the insulin protein. Explain.
16. Based on what you observed, which bonding interactions occur first, hydrogen bonds between beta sheets or disulfide bonds? Why?
